Note
Go to the end to download the full example code.
Invert Uncertain Transform#
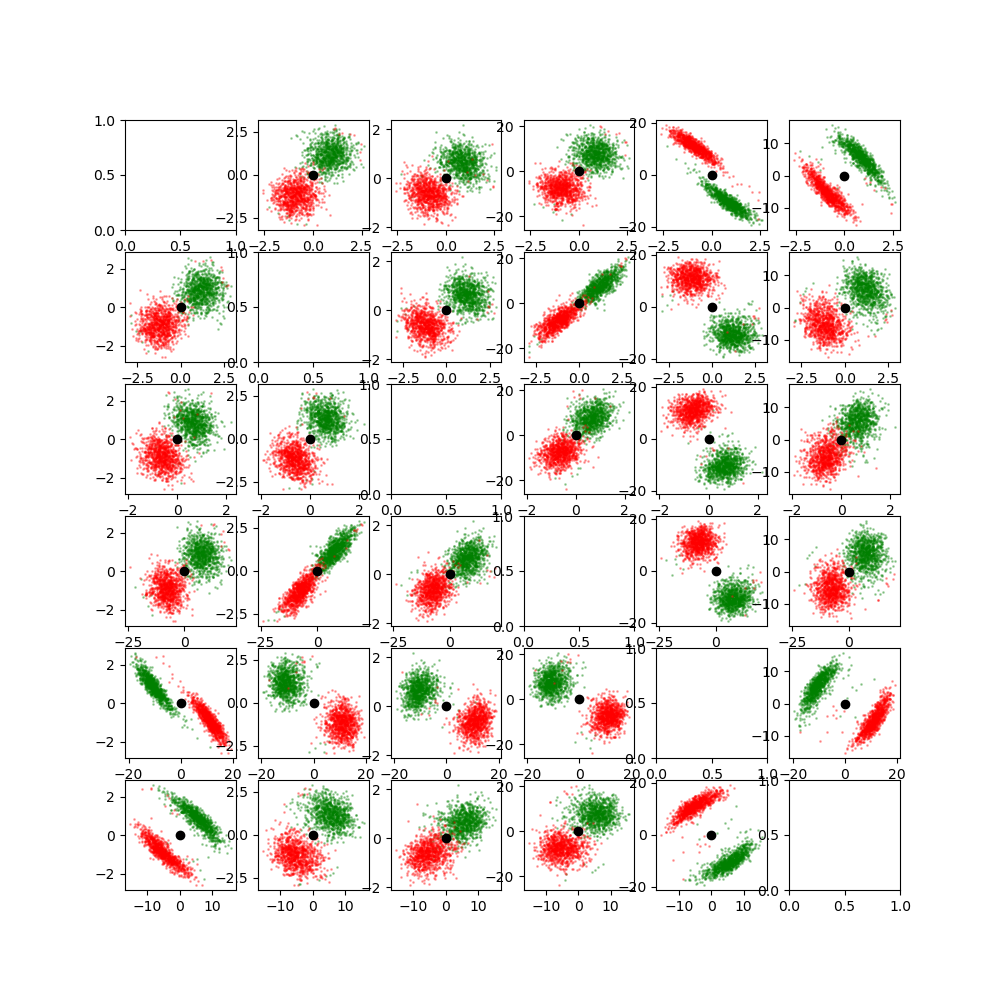
We sample from the original transform distribution and from the inverse distribution. Samples are then projected to all 2D planes and plotted. The color indicates the pose distribution. Green is the original distribution and red is the inverse.
import matplotlib.pyplot as plt
import numpy as np
import pytransform3d.rotations as pr
import pytransform3d.trajectories as ptr
import pytransform3d.transformations as pt
import pytransform3d.uncertainty as pu
n_mc_samples = 1000
rng = np.random.default_rng(1)
alpha = 2.0
A2B = pt.transform_from(
R=pr.matrix_from_euler([1.5, 0.5, 1.3], 0, 1, 2, True), p=[10.0, -7.0, -5.0]
)
variances = alpha * np.array([0.1, 0.2, 0.1, 2.0, 1.0, 1.0])
std_devs = np.sqrt(variances)
correlations = np.array(
[
[0.5, 0, 0, 0, 0, 0],
[0.1, 0.5, 0, 0, 0, 0],
[-0.3, 0.1, 0.5, 0, 0, 0],
[-0.1, 0.2, 0.3, 0.5, 0, 0],
[-0.3, -0.1, -0.2, 0.3, 0.5, 0],
[-0.1, 0.1, 0.2, 0.1, 0.3, 0.5],
]
)
correlations += correlations.T
cov_A2B = correlations * np.outer(std_devs, std_devs)
x_A2B = pt.exponential_coordinates_from_transform(A2B)
samples_A2B = ptr.exponential_coordinates_from_transforms(
[pt.random_transform(rng, A2B, cov_A2B) for _ in range(n_mc_samples)]
)
B2A, cov_B2A = pu.invert_uncertain_transform(A2B, cov_A2B)
x_B2A = pt.exponential_coordinates_from_transform(B2A)
samples_B2A = ptr.exponential_coordinates_from_transforms(
[pt.random_transform(rng, B2A, cov_B2A) for _ in range(n_mc_samples)]
)
_, axes = plt.subplots(nrows=6, ncols=6, squeeze=True, figsize=(10, 10))
for i in range(6):
for j in range(6):
if i == j:
continue
indices = np.array([i, j])
ax = axes[i][j]
ax.scatter(
samples_A2B[:, i], samples_A2B[:, j], color="g", s=1, alpha=0.3
)
ax.scatter(
samples_B2A[:, i], samples_B2A[:, j], color="r", s=1, alpha=0.3
)
ax.scatter(0, 0, color="k")
plt.show()
Total running time of the script: (0 minutes 1.333 seconds)